Our laboratory currently focuses on following research themes:
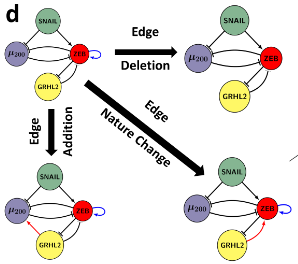
Dynamics of Epithelial-Mesenchymal Plasticity
Epithelial-mesenchymal plasticity (EMP) involves reversible and dynamic cell-state transitions among epithelial, mesenchymal and hybrid epithelial/mesenchymal (E/M) phenotypes. We focus on identifying the molecular factors stabilizing hybrid E/M cell-states, and characterizing the association of EMP with other axes of plasticity, such as tumor-initiation potential, immune evasion etc.
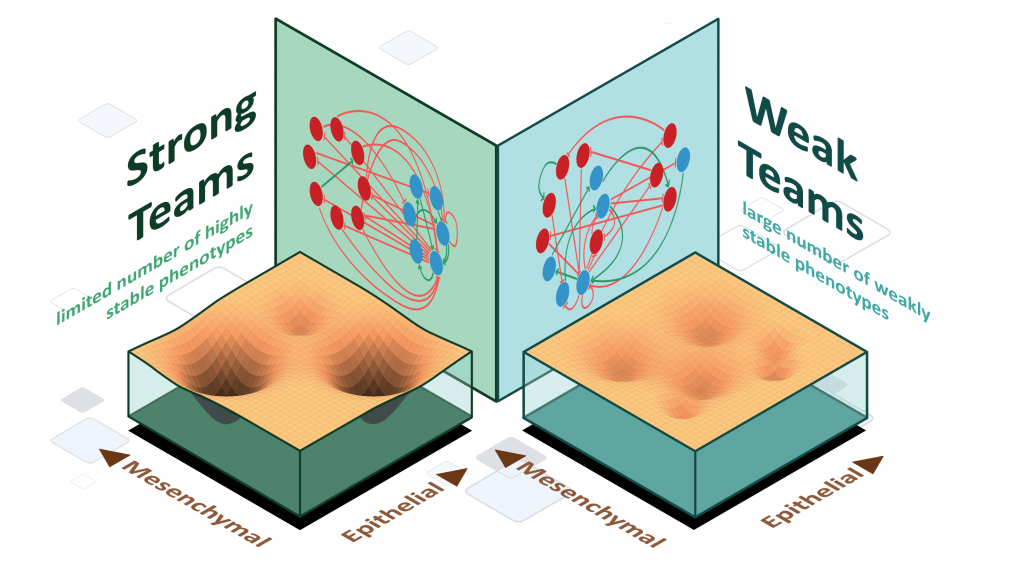
Mechanisms underlying phenotypic heterogeneity in cell populations
Stochastic cell-state transitions driven by various factors such as epigenetic modifications and asymmetric cell division can enable phenotypic heterogeneity in a genetically identical population. Such heterogeneity can enable bet-hedging, thus facilitating survival of a population under stress and the relapse of population upon stress withdrawal. We aim to identify different mechanisms and underlying principles enabling such phenotypic heterogeneity in diverse biological contexts.
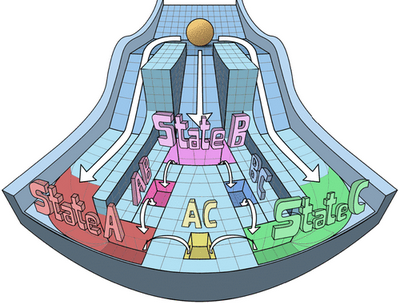
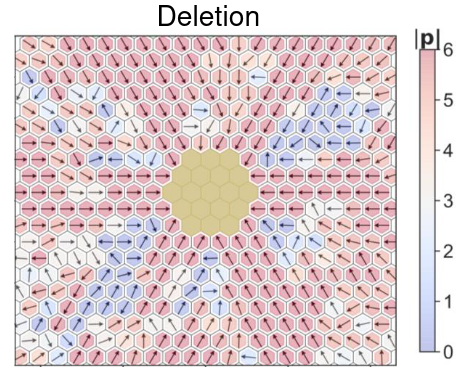
Spatiotemporal pattern formation and collective behavior in cell populations
Spatiotemporal dynamics of cellular phenotypes are often driven by their crosstalk with their microenvironment, involving cell-cell communication based feedback loops. We develop multi-scale mechanism-based models as predictive ‘in silico co-culture’ systems to elucidate the collective emergent dynamics in multiple scenarios: tissue-level patterning, collective cell migration, and tumor-stroma crosstalk.
Single-cell analysis of cell-fate decisions in development and cancer
Recent single-cell high-throughput data collection has enabled tracking the trajectories of cell-fate decisions and mapping phenotypic heterogeneity in diverse biological contexts. We examine such single-cell transcriptomic data to identify branching points in decision-making, to characterize subpopulations with specific molecular and functional attributes, and to probe the dynamic evolution of different modules of co-expressed gene programs.
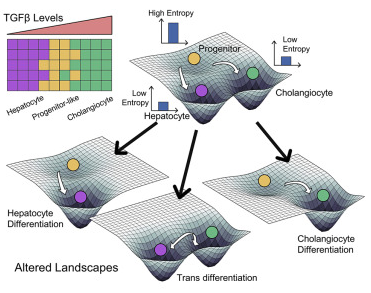
Non-genetic mechanisms of adaptive therapy resistance
Besides genetic mutations, non-genetic mechanisms such as phenotypic switching and stochastic cell-to-cell heterogeneity can enable adaptation to various stresses. For instance, drug treatment can induce a phenotypic switch in a fraction of cells to another state (Lamarckian induction). We aim to elucidate how cells adapt reversibly to various stresses, and how we can design strategies to outcompete these adaptation strategies.
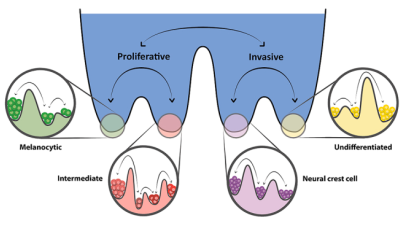
Design principles of cell-fate decision-making regulatory networks
Cell-fate decision-making regulatory networks often have unique features in terms of their emergent dynamics, for instance, a limited number of phenotypes despite their complex and large architecture, and balancing robustness against various perturbations with enabling cell-state switching. We are interested in decoding the topological hallmarks of biological networks, and the functional implications of those hallmarks.